library(INLAvaan)
mod <- "
# Latent variable definitions
ind60 =~ x1 + x2 + x3
dem60 =~ y1 + y2 + y3 + y4
dem65 =~ y5 + y6 + y7 + y8
# Latent regressions
dem60 ~ ind60
dem65 ~ ind60 + dem60
# Residual correlations
y1 ~~ y5
y2 ~~ y4 + y6
y3 ~~ y7
y4 ~~ y8
y6 ~~ y8
"
dat <- lavaan::PoliticalDemocracy
# Simulate missingness (MCAR)
set.seed(221)
mis <- matrix(rbinom(prod(dim(dat)), 1, 0.99), nrow(dat), ncol(dat))
datmiss <- dat * mis
datmiss[datmiss == 0] <- NAINLAvaan handles missing data in one of two ways: listwise deletion (default, i.e. uses all complete cases) or Full Information Maximum Likelihood (FIML; missing = "ML").
Simulate Data
Listwise Deletion
fit1 <- asem(mod, datmiss, meanstructure = TRUE)
#> ℹ Finding posterior mode.
#> ✔ Finding posterior mode. [156ms]
#>
#> ℹ Computing the Hessian.
#> ✔ Computing the Hessian. [419ms]
#>
#> ℹ Performing VB correction.
#> ✔ VB correction; mean |δ| = 0.076σ. [417ms]
#>
#> ⠙ Fitting skew normal to 0/42 marginals.
#> ⠹ Fitting skew normal to 12/42 marginals.
#> ✔ Fitting skew normal to 42/42 marginals. [3.2s]
#>
#> ℹ Sampling covariances and defined parameters.
#> ✔ Sampling covariances and defined parameters. [197ms]
#>
#> ⠙ Computing ppp and DIC.
#> ⠹ Computing ppp and DIC.
#> ✔ Computing ppp and DIC. [630ms]
#>
fit1@Data@nobs[[1]] == nrow(datmiss[complete.cases(datmiss), ])
#> [1] TRUE
print(fit1)
#> INLAvaan 0.2.3.9004 ended normally after 71 iterations
#>
#> Estimator BAYES
#> Optimization method NLMINB
#> Number of model parameters 42
#>
#> Used Total
#> Number of observations 35 75
#>
#> Model Test (User Model):
#>
#> Marginal log-likelihood -818.357
#> PPP (Chi-square) 0.102
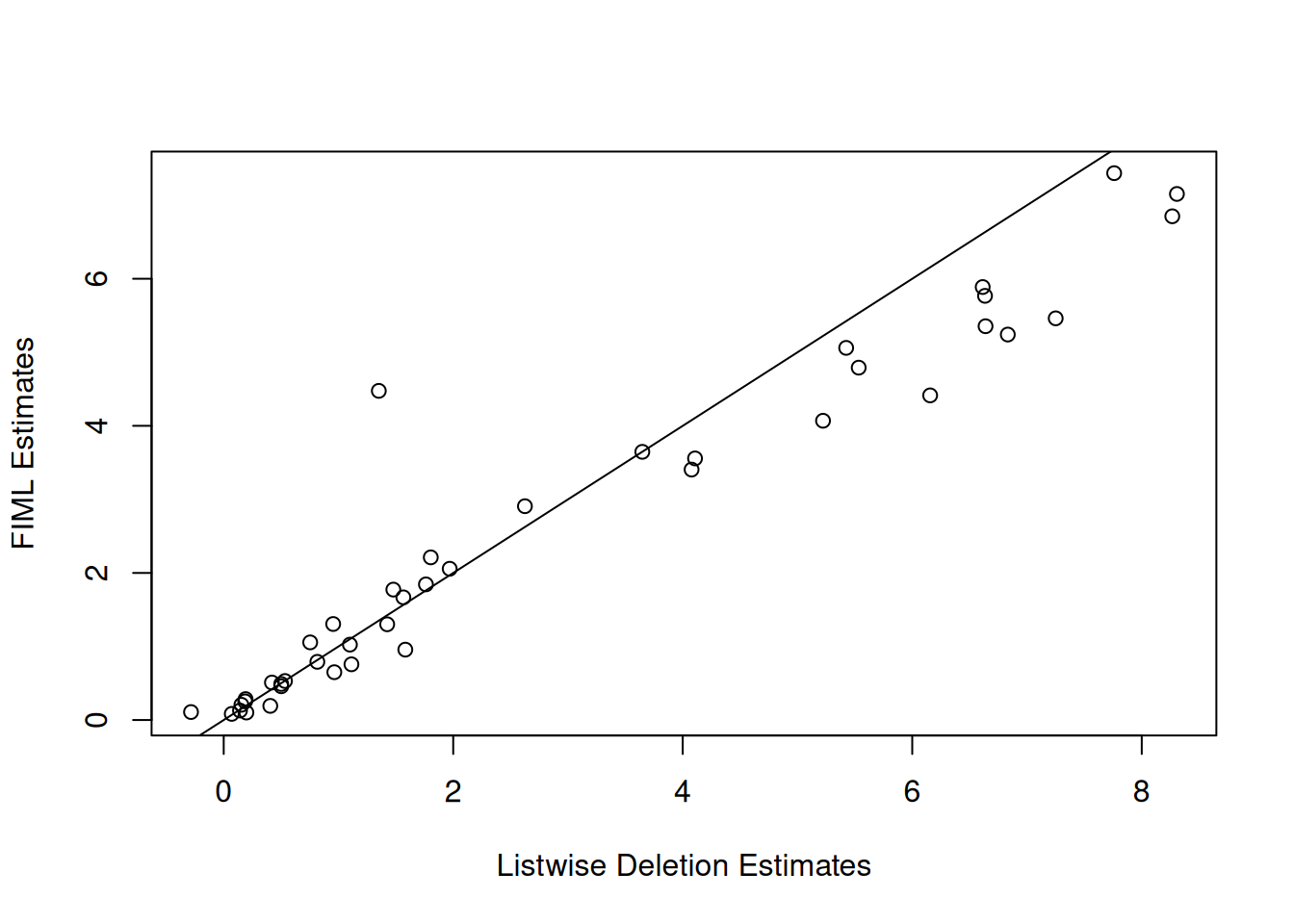
coef(fit1)
#> ind60=~x2 ind60=~x3 dem60=~y2 dem60=~y3 dem60=~y4 dem65=~y6
#> 1.804 1.762 0.964 0.816 1.583 1.100
#> dem65=~y7 dem65=~y8 dem60~ind60 dem65~ind60 dem65~dem60 y1~~y5
#> 0.754 1.425 0.954 0.534 1.113 0.187
#> y2~~y4 y2~~y6 y3~~y7 y4~~y8 y6~~y8 x1~~x1
#> 0.406 0.501 0.198 -0.285 0.191 0.070
#> x2~~x2 x3~~x3 y1~~y1 y2~~y2 y3~~y3 y4~~y4
#> 0.142 0.420 1.566 7.760 4.077 2.625
#> y5~~y5 y6~~y6 y7~~y7 y8~~y8 ind60~~ind60 dem60~~dem60
#> 1.478 6.614 1.969 3.648 0.502 1.352
#> dem65~~dem65 x1~1 x2~1 x3~1 y1~1 y2~1
#> 0.154 5.424 5.534 4.107 7.250 6.634
#> y3~1 y4~1 y5~1 y6~1 y7~1 y8~1
#> 8.306 6.833 6.639 5.223 8.266 6.156Full Information Maximum Likelihood (FIML)
fit2 <- asem(mod, datmiss, missing = "ML", meanstructure = TRUE)
#> ℹ Finding posterior mode.
#> ✔ Finding posterior mode. [258ms]
#>
#> ℹ Computing the Hessian.
#> ✔ Computing the Hessian. [787ms]
#>
#> ℹ Performing VB correction.
#> ✔ VB correction; mean |δ| = 0.047σ. [521ms]
#>
#> ⠙ Fitting skew normal to 0/42 marginals.
#> ⠹ Fitting skew normal to 8/42 marginals.
#> ⠸ Fitting skew normal to 30/42 marginals.
#> ✔ Fitting skew normal to 42/42 marginals. [5.8s]
#>
#> ℹ Sampling covariances and defined parameters.
#> ✔ Sampling covariances and defined parameters. [200ms]
#>
#> ⠙ Computing ppp and DIC.
#> ✔ Computing ppp and DIC. [885ms]
#>
print(fit2)
#> INLAvaan 0.2.3.9004 ended normally after 93 iterations
#>
#> Estimator BAYES
#> Optimization method NLMINB
#> Number of model parameters 42
#>
#> Number of observations 75
#> Number of missing patterns 19
#>
#> Model Test (User Model):
#>
#> Marginal log-likelihood -1414.932
#> PPP (Chi-square) 0.000
coef(fit2)
#> ind60=~x2 ind60=~x3 dem60=~y2 dem60=~y3 dem60=~y4 dem65=~y6
#> 2.211 1.844 0.650 0.791 0.957 1.025
#> dem65=~y7 dem65=~y8 dem60~ind60 dem65~ind60 dem65~dem60 y1~~y5
#> 1.055 1.301 1.305 0.530 0.757 0.253
#> y2~~y4 y2~~y6 y3~~y7 y4~~y8 y6~~y8 x1~~x1
#> 0.192 0.490 0.101 0.108 0.284 0.083
#> x2~~x2 x3~~x3 y1~~y1 y2~~y2 y3~~y3 y4~~y4
#> 0.129 0.510 1.668 7.436 3.405 2.907
#> y5~~y5 y6~~y6 y7~~y7 y8~~y8 ind60~~ind60 dem60~~dem60
#> 1.773 5.887 2.057 3.646 0.459 4.476
#> dem65~~dem65 x1~1 x2~1 x3~1 y1~1 y2~1
#> 0.207 5.060 4.791 3.557 5.462 5.768
#> y3~1 y4~1 y5~1 y6~1 y7~1 y8~1
#> 7.153 5.241 5.354 4.069 6.849 4.414